How to use Genesys
August 15, 2023
A collection of step-by-step guides and webinars on all things Genesys.
- Returning users: catch up with the 2022 user interface updates
- Get familiar with the functionalities of Genesys
- Search for accessions
- Search for particular traits
- Search for accessions by their climate at origin
- Create your own subsets
- Order accessions from genebanks through Genesys
- Watch video tutorials in English
- Watch video tutorials in French
1. Returning users: catch up with the 2022 user interface updates
If you were using Genesys before mid-2022, you are probably familiar with this navigation menu.

Well, it looks a bit different now.

Yes, the navigation menu bar was remodeled, and now includes the following items:
-
Accession data collects the pages of all four data types in Genesys: passport data, subsets, characterization and evaluation (C&E) datasets, and descriptor lists.
-
Directory clusters the passport data in different pages, either by crop, geography, data provider, or network.
-
Resources includes all reading material, from the About section to the Recent Activity blog to the documentation pages.
-
My List remains the same.
New in 2022: Table view for passport data
On the Accession browser page you can now toggle between a List view and a Table view. The original List view displays only the key passport data of the accessions listed. The new Table view shows the passport data in a tabular view (as expected), which allows for easier spotting of accessions of interest. The Table view will soon become the default setting for all users.

In this view, selecting accessions to add or remove from My List is available under the floating action button in the bottom right corner. First, select accessions by clicking or tapping the cell with row numbers; then, select Add to My list or Remove from My list.

You can filter out descriptors that you do not wish to visualize in the Table view by clicking or tapping the settings icon (⚙️) on the right-hand side and turning off (🔘) the descriptors you wish to hide, then selecting the save icon (💾) to close the table configuration panel.


If you have any suggestions for further user interface or user experience updates, please feel free to contact helpdesk@genesys-pgr.org.
2. Get familiar with the functionalities of Genesys
Genesys for genebanks
The first page of this handout summarizes all the functionalities of Genesys that are most interesting for genebanks. It provides an overview of the benefits for your genebank when you join and start sharing data in Genesys.
Sharing accession data on Genesys can help increase the visibility of your genebank’s collection, promote your work, and make the data more accessible to researchers and breeders. Genesys provides tools to help you manage and organize your data, and offers Embedded Genesys to easily integrate data directly into your existing websites. We also promote the work of genebanks that publish data in Genesys through blog posts and newsletters that reach a global audience.
Genesys is a vital source of data for conducting gap analyses and tracking the current conservation status of crops. Data in Genesys is used by FAO for their reporting on the Sustainable Development Goals. As an important part of the Global Information System of the International Treaty on PGRFA, Genesys is connected to other key databases, such as FAO WIEWS, GRIN Taxonomy for Plants, Easy-SMTA, and the DOI Registration Service.
Genesys for users of genebanks and their data
For users of genebanks and their data, Genesys is your one-stop-shop for the crop diversity conserved around the world. The second page of this handout lists all the tools available in Genesys to search through more than four million accessions.
Genesys is a powerful tool for finding the specific crop diversity you need for research or breeding. The data in Genesys is collated using international standards for exchange of crop germplasm information, and includes over 150,000 images, 400 characterization and evaluation datasets, and 250 core collections and other interesting subsets of plant genetic resources. You can download the data to conduct your own analyses for your specific purposes.
Once you have found the crop diversity you need, you can submit a request for the germplasm to most of the participating genebanks directly through Genesys, or reach out to genebank curators for assistance. We cannot guarantee you access to the material, since that is subject to international and national law, and the policies of each genebank. But you have to start somewhere…
Join us!
If you are a genebank looking into joining Genesys, send us an email at helpdesk@genesys-pgr.org.
If you are a user of genebanks looking into making use of the tools Genesys has to offer, make sure you register an account at https://www.genesys-pgr.org/registration.
3. Search for accessions
There are many ways to find the accessions you are looking for in Genesys. On the Genesys homepage, you can begin your search for accessions, subsets, datasets, or descriptor lists by entering the keywords of interest.

The text search is a useful way to get started with Genesys, but it is not the silver bullet you may be hoping for. The most reliable method to query Genesys is by using the data filters described in this tutorial.
Layout
Regardless of how you access the accession passport data page, the filters pane will be on the left-hand side, with the filters organized into groups (which are collapsed by default). The main section of the layout presents accession data in a few different ways using the five tabs at the top.

The Overview tab summarizes the number of accessions matching your filters by different categories. This tab provides an overall picture of accession data. The second tab, Accessions, shows passport data row by row. This is also where you can download the passport data of your selected accessions. Map displays the localities where the material was collected (matching any filters you have applied). Images provides access to a gallery of pictures associated with accessions (again matching your filters). The final tab, Subsetting Tool, lets you create subsets when you are logged into a Genesys account.
The content shown in this main section of the layout always corresponds to urrenttly applied filters. A good understanding of which filters to apply, and how, will enable you to find accessions of interest faster and more reliably.
Filter groups
- Historical records
- Text search
- Holding institute
- Accession number
- Date search
- Crop
- Taxonomy
- Origin of material
- Collecting data
- Biological status of accession
- Type of germplasm storage
- Status
- Referenced accessions
- Climate at origin
Historical records
A historical record is the passport data of an accession that no longer exists in the record-keeping genebank collection. By default, historical records are excluded from the search.
Text search
Whether you type in a species name, country, or simply a number, the full text search will scan all the passport data in Genesys and yield accessions with the associated text.
You can build a search query using double quotes to limit the search to an exact phrase (“rice”); a pipe symbol as an OR operator (rice | leaf); an asterisk to match a prefix (rice*); and parentheses for grouping.
You can even search accessions for words that may be spelled slightly differently using a fuzzy search. Apply a fuzzy search by adding a tilde at the end of a word (solanum~).
💡 Tip: A full text search query will not necessarily result in a 100% match. Other filters below may offer greater precision.
Holding institute
A holding institute is the genebank that conserves an accession. International, regional, and national genebanks are the primary contributors of data to Genesys. They are all listed in our data providers directory, either as independent partners or under data providers, since some of our partners aggregate and provide data to Genesys on behalf of several genebanks.
Institute code
You can narrow your search to only accessions in a specific genebank using the genebank’s WIEWS institute code. Type the first few letters of the genebank’s full name or acronym and Genesys will auto-complete your typing. Note that only the first 10 matches are displayed, sorted by the number of each institute’s accessions in Genesys. You may need to enter more characters to find the institute you are looking for.

If you have not entered anything in the field, Genesys simply displays the top 10 institutes by number of accessions. You can apply one of these suggested options by clicking or tapping on it. The list of top 10 suggestions is automatically updated as you add other filters to your search query. If you filter for Zea mays, for example, the top 10 suggestions will include only the institutes with Zea mays accessions.
Country of holding institute
Instead of focusing on one or more institutes, you may want to focus your search on the country where the genebank is located, i.e. the country of the holding institute. For example, to filter for accessions that are conserved in genebanks in Italy, type in the country name to bring up the auto-completer, or enter the country’s ISO code (ITA).
Accession number
The accession number is the unique identifier of the material in the collection. This identifier is often composed of a prefix, a number, and a suffix (e.g. “Tme-419”, “AGG 5 WHEA”).
This filter allows you to search for accessions by entering accession numbers manually, by copy-pasting a list of accession numbers from a spreadsheet, or by specifying the minimum and maximum sequential number.

Press the Enter key to add more accession numbers to the query.

💡 Tip: You can copy a list of accession numbers from Excel and paste it into the Accession number filter.
Sequential number
Genesys automatically extracts the numerical part of the accession number so that you can query for a numeric range of accession numbers.
Date search
There are many dates associated with accessions, such as when they were acquired by the genebank or when they were collected from the field. The date search filters are concerned specifically with metadata about the accession information. There are three filters:
Created on
This filters by the time period when the accession information was created in Genesys for the first time.
Last modified
This filters by the time period when the accession information was most recently updated or modified in Genesys.
Taxonomy last modified
This filters more specifically by the time period when the taxonomy of an accession was most recently updated or modified.

The date search filters always work in a range, with the left-hand date meaning “created/last modified starting...” and the right-hand date meaning “created/last modified before...”. When either is left unspecified, this indicates “whenever”. If you need to filter for one specific day, then you need to select this date in both the left and right field.

Crop
Genesys aggregates accession data based on the crop names in English as provided by genebanks, and enables you to search these aggregations by crop.
💡 Tip: Crop aggregations are very limited and may be incomplete due to differences in spelling or local names of crops. For a more accurate search and overview of the accessions in Genesys, search by taxonomic names instead.
Taxonomy
You can filter out accessions by their scientific names in different ways:
-
Genus name (make sure to capitalize the first letter, for example: Zea)
-
Species, meaning both the genus name and species name (for example: Zea mays)
-
Specific epithet, meaning only the species name (for example: mays)
-
Subtaxon, any level of taxonomy underneath the species name
GRIN taxon ID
The four fields above allow you to filter for accessions by their taxonomy as provided by the genebank. Alternatively, you may filter for accessions according to the GRIN Taxonomy. The Genesys algorithm determines this by trying to match the genebank’s taxonomy with the GRIN Taxonomy, and allows users to filter by the resulting GRIN Taxon ID.
💡 Tip: you can copy and paste a list of cells from Excel in this filter.
Matching to GRIN Taxonomy is also what allows Genesys to include synonyms in the search results using taxonomy. Look for the button “Include synonyms in search results” under the Taxonomy filter group.

Origin of material
Provenance of material filters passport data according to the country from which the material comes. If an accession was collected in the field during the course of a collecting expedition, then its provenance is the country of the collecting site. If the material is a breeding line, then its provenance is the country where the material was developed.
Alternatively, you may search for an accession by its origin at specific geographic coordinates such as elevation, longitude, and latitude (in decimal format). These filters work using ranges of numerical values. To filter for accessions where the elevation is over 1,000 meters, enter 1000 in the left field, At least (min). To filter for accessions where the elevation is at most 2,000 meters, enter 2000 in the right field, At most (max). Using both of these will filter for accessions where the elevation is between 1,000 and 2,000 meters.

💡 Tip: If you want to look for a specific number instead of a range, you need to insert the value in both the minimum and maximum fields.
Collecting data

This filter group enables you to filter by information related to the accession collecting mission. It includes the following filters:
Collecting date
This is the collecting date of the accession in YYYYMMDD format, where YYYY is the year, MM is the month, and DD is the day. If you do not wish to specify a month or day, you can replace the MM or DD with two hyphens or zeroes (-- or 00).
Collecting number
This is an original identifier assigned by the collector(s) of the sample, normally composed of the name or initials of the collector(s) followed by a number (e.g. “FM9909”).
Collecting mission
This is the identifier of the collecting mission used by the collecting institute (e.g. “CIATFOR-052”, “CN426”).
Location of collecting site
This is location information below the country level that describes where the accession was collected. This might include the distance in kilometers and direction from the nearest town, village or map grid reference point (e.g. “7 km south of Curitiba in the state of Parana”).
Biological status of accession
Here you can filter for accessions by their level of improvement: wild, landrace, breeding material, etc.

Type of germplasm storage
Ex situ genebanks maintain plant genetic resources as seed, in the field, in vitro, in cryo, or in DNA collections. A single accession may be maintained in different conditions; therefore, multiple options are possible.
Status
This filter category groups six variables about an accession related to its status in Genesys, in the genebank, and in relation to different international bodies.
Available for distribution
Through this status variable, the genebank indicates to Genesys users whether the accession has enough inventory to be distributed.
Georeferenced, with coordinate data
This is a simple filter to limit the search to accessions with (or without) geographic coordinate data (latitude and longitude).
Included in the MLS
Through this variable, the genebank indicates whether the accession is in the Multilateral System (MLS) of the International Treaty on Plant Genetic Resources for Food and Agriculture.
Backed up in SGSV
Through this variable, the genebank indicates whether the accession is backed up in the Svalbard Global Seed Vault (SGSV).
Accession with images
This will limit the search to accessions with (or without) images of the accession published in Genesys.
AEGIS accession
This filters by whether the accession is part of A European Genebank Integrated System (AEGIS), an initiative by the European Cooperative Programme for Plant Genetic Resources aiming to efficiently conserve and provide access to unique germplasm in Europe through the establishment of the European Collection.
Referenced accessions
Many accessions are included in the subsets or datasets that genebanks publish in Genesys. In order to filter out accessions that are part of a certain subset or dataset, copy the unique ID from the set’s page in the Data and resources section (see the highlighted part in the screenshot below).

Then paste this unique ID in the passport data filters sidebar. For one dataset, you can also click or tap Filter Accessions on the dataset page directly – but copying and pasting the unique ID allows for filtering accessions from many sets simultaneously.

💡 Tip: Check out at the end of this page the recording of our webinar on subsets and trait data in Genesys for more information.
Climate at origin
Genesys allows you to filter for accessions according to 19 temperature and precipitation variables from collecting sites. This filter only works for accessions with geographic coordinates provided by the genebanks.
A separate tutorial below details how to use this filter.
Tips and tricks
Directory pages
In the Directory navigation menu on the top banner of Genesys, you will find pages that aggregate accessions by country of provenance, crop, and holding institute. These pages include filtered accessions based on those categories.
Getting an overview as you filter
The filters sidebar also acts as a mini-dashboard where you can get an overview of the distribution of accessions across the different variables – for example, how many accessions in the current filtering status are aggregated under the amaranth crop.

After filtering accessions using any option, not only do the overview numbers re-adjust, but also the filter choices themselves. For example, if you filter for accessions collected from Uruguay and none of the results are tagged as amaranth, then this option will be rendered invisible under the Crops filter group. This is one of the many ways Genesys makes it convenient to sift through over 4 million records.
Excluding matches
Genesys will search for the value you provide for a filter, and these values are prefixed by a plus symbol. For example, filtering for rice accessions shows that many are maintained by the International Rice Research Institute genebank (PHL001) – but you may want to exclude IRRI accessions from the search. Just filter by this Institute code, and click or tap the “+” to toggle it to “-”. You are now instructing Genesys that the results must exclude the specified value.

The excluded PHL001 is displayed at the top of the page when this change is applied.

💡 Tip: The number of accessions that you see at the top of the page after filtering sometimes has an “about” before it, as here. This means that the number is an approximation; Genesys does this to increase the performance of the software. You can get the exact number by downloading the MCPD file of the passport data in your results.
Removing individual filters
The top of the page is your go-to in order to remove one of the filters you applied without resetting them all. In the screenshot above, if you changed your mind about filtering by the Oryza genus, you could simply click or tap the “x” button of that filter.
Further examples and help
In case you missed it, there is a recording of the webinar we organized on how to use Genesys that includes examples of how to use these filters, and more!
If you have any questions, send them over to helpdesk@genesys-pgr.org.
4. Search for particular traits
Characterization and evaluation (C&E) data stimulates the use of germplasm collections. Since 2018, Genesys has provided access to C&E datasets from various genebanks for download and analysis by users. But querying trait data directly in Genesys was not possible until the 2022 update of Genesys. With this update, it became possible to search the C&E datasets for specific traits.
Searchable datasets display the following notice, and you can access them by clicking or tapping the banner.

Searchable datasets follow the familiar Genesys layout: the filtering options are listed on the left, and the main display area is used to show trait data that match the specified filters. The data are initially not filtered and the grid allows you to browse through the entire dataset.

The Observations tab
The Observations tab displays trait data in a grid, one row per accession.
The first column (My List) indicates whether the accession is already in your list. Tick the checkbox to add it, and untick the checkbox to remove it.
The Accession column displays the accession number with a link to its passport data.
The remaining columns are for the traits included in the dataset. Multiple observations may be listed for individual traits, and when searching for traits of interest, a match is made if any of the observations of the trait fit the criteria.
The initial order of traits in the grid is determined by the genebank when it registers the dataset in Genesys. As with all other grids in Genesys, you can rearrange the columns by dragging them to the desired position.
The settings icon (⚙️) on the right edge of the grid header allows you to toggle the visibility of columns and adjust their display: wrap text, apply heavier font weight, and adjust text alignment. You can close the grid settings by clicking or tapping on the save icon (💾).

Grid settings are saved in your browser and will be used every time you visit this dataset.
Downloading grid data
The action button in the bottom right corner of the grid allows you to download the data in Excel format. It downloads all data that matches your current filters, not only the rows currently loaded in the grid.

Filtering trait data
The left panel lists the individual traits: their names, descriptions, filtering options, and a link to the definition of the descriptor that was used. The order of filters follows the order of traits when the dataset was registered.
Crop descriptors document how the observations were recorded. The filtering options depend on the type of descriptor.
The most common type of descriptor in characterization data is coded. These are used to capture categorical data and specify what each code represents.
Filtering options for coded descriptors are displayed as a list of checkboxes, indicating that multiple options can be selected.

Quantitative data from exact measurements (counts, lengths, widths, etc.) or their mean values use numerical descriptors. In addition to the methodology used to make the observations, these must specify the unit of measurement.
Numerical descriptors allow you to enter a value range, where At least means the observed value must be greater than or equal to this value (i.e. the minimum value) and At most requires that the trait value be less than or equal to the specified maximum.

The minimum or maximum value can be left blank for the total range.
Numerical observations are sometimes normalized and expressed on a scale, for example from short to tall, or from susceptible to resistant. Such ordinal scale descriptors are often found in characterization and evaluation data. In the example below, the damaged area of leaves is not expressed as the exact percentage of leaf area, but as a scale where leaf damage goes from low damage (1) to extensive damage (5). You can adjust the range of interest by dragging the two circular buttons.

Text descriptors are used for unstructured data such as notes and comments. Entering a value in a text filter will match accessions where the note contains the phrase specified in the filter (e.g. “2006” will match both “2006DS” and “DS2006”).

Applying the filters
Genesys does not automatically refresh the data in the grid when you set the trait filters. You must click or tap Apply filters for the filters to work.
Your combination of filters may result in zero matching records. If that happens, try relaxing the criteria by specifying a wider option or by removing one or more filters.

The Overview tab
After Observations, the next tabs are Overview, Analyze (this is a work in progress, and we’ll reserve it for later), and Description.
The Overview tab provides various visualizations of the data. Numerical traits are shown as histograms, while coded and scale descriptors use bar charts.
These visualizations reflect the selected data range (records that match your filters) and may help you in determining which filters to apply next!

The Analyze tab
This tab shows heatmaps of relationships between categorical data (descriptors using a fixed vocabulary of options).
This is still work in progress and subject to change.
The Description tab
This tab displays the metadata for the dataset. It also has links to download the source files as uploaded by the genebank, including trait observations and the passport data of accessions included in the dataset.

Some history
In 2018, we completed the “Genesys Catalog of Phenotypic Datasets Linked to Genebank Accessions” project, which aimed to make accession-level data on the plant genetic resources that have been characterized or evaluated by genebanks available to researchers. The work was supported by the Federal Republic of Germany (project reference GenR 2016-1).
We learned that requiring data to be provided in a specific format results in reluctance to share data, primarily because genebanks were not able to reliably convert the data from its original shape to the required standard format. The project therefore focused on providing access to data files as produced by genebanks, without any interpretation that might cause loss of information.
Genebank C&E datasets come in all shapes and sizes, making the unpacking and interpretation of data quite a complex task. Genesys enables genebanks to validate and document trait data as part of the publishing process. They provide assurance that data is correctly decoded and linked to a descriptor that fully documents the trait and its encoding.
What is next?
If the Catalog project was the first big step in making trait data more easily available in Genesys, then making them searchable marks the second step. The visualizations and searching should make it more attractive for genebanks to publish datasets in Genesys. And of course more attractive for users to explore the data and hopefully easier to find the diversity they want.
As genebanks often use a fairly consistent set of descriptors for their accessions of a particular crop, it may be possible to make traits searchable across multiple datasets of one crop in a given genebank. Making data searchable across genebanks is a bigger challenge, as genebanks unfortunately do not all use the same descriptors. When traits “Grain color” or “Susceptibility to rust” are coded differently, the descriptors first need to be mapped and their data made compatible for searching. This will require development of methods to make different descriptors somehow compatible.
We’re on it.
5. Search for accessions by their climate at origin
Genesys makes it possible to search across more than 4 million accessions, and of these, more than 1 million are accompanied by the geographical coordinates of the sites where the material was originally collected. You will see such georeferenced accessions included in the maps generated by Genesys.
This tutorial documents some ways you can use climatic parameters to find matching georeferenced accessions in Genesys. Dr. Alan Humphries, curator at the Australian Pastures Genebank, has also compiled a comprehensive video guide on this type of searching.
How searching by climate at origin works
If we assume that plants adapt to their environment with time, then accessions collected from warmer environments, for example, should be more tolerant to hot conditions compared to those collected from areas with cooler climates. This is useful information for many genebank users.
Genesys links the coordinates of georeferenced accessions with data about the climate at the corresponding location. The climate data is sourced from WorldClim, a global climate dataset with a spatial resolution of about 1 km2 at the equator. WorldClim includes a number of bioclimatic variables useful for our purposes:
Temperature variables
-
Annual: Annual mean temperature and annual temperature range in °C
-
Quarterly: Mean temperatures of wettest, driest, warmest, and coldest quarters in °C
-
Seasonal: Temperature seasonality (standard deviation *100)
-
Monthly: Maximum temperature of warmest month and minimum temperature of coldest month in °C
-
Mean diurnal temperature range (mean of monthly (max temp – min temp))
-
Isothermality
Precipitation variables
-
Annual: Annual precipitation in mm
-
Quarterly: Precipitation of wettest, driest, warmest, and coldest quarter in mm
-
Seasonal: Precipitation seasonality
-
Monthly: Precipitation of wettest month and driest month in mm
Selecting a point on the map
The simplest way to choose the climate for which you wish to find accessions is by simply selecting any land area on the map in Genesys. This will display the climate at the selected location and offer the option to list the accessions that are linked to similar temperature and precipitation conditions.

Choosing the climate based on an accession
Alternatively, for any accession that includes coordinates, you can ask Genesys to display the climatic conditions at the accession’s origin and find accessions that match the temperature and precipitation conditions of that location. You can get a list of these matches using this button in an accession’s details page.

Adjusting the filters sidebar
The two options described above only use a carefully selected set of climatic filters: annual mean temperature, seasonality, and rainfall. You can adjust or specify your own filters in the Climate at origin section, which is at the bottom of the filters sidebar, after all the taxonomy and provenance filters.

The precipitation and temperature variables, for which you can set the minimum and/or maximum values, are grouped under the Climate at origin section.
Making use of the results of a climatic search
If you want to order samples of any of the accessions identified by these searches from the genebanks that conserve them, check out the below tutorial on ordering accessions from genebanks through Genesys.
You may also want to download the passport data or KML file of these accessions for your own analysis. In this case, you will need to log in to your Genesys account and use the Download MCPD or Download KML buttons.

6. Create your own subsets
Genesys includes many subsets created by genebank experts to help users find particular kinds of interesting material within larger collections.
You can now create your own subsets of accessions, using the climate and soil variables of your choice, in the Subsetting Tool.
The Subsetting Tool, developed by the Alliance of Bioversity International & CIAT and the International Center for Agricultural Research in the Dry Areas (ICARDA), allows Genesys users to use combinations of abiotic factors to create subsets of accessions that can grow under specific environmental conditions. Think of it as a do-it-yourself method for subsets with particular adaptations.
The tool only considers accessions that are published in Genesys with the coordinates of their collecting site in the passport data. With this data, you can use 20 agroclimatic indicators related to the site of collecting to do the subsetting, grouped into six categories: drought, flooding, heat, photoperiod, soil, and in some cases additional crop-specific indicators.
How the Subsetting Tool works
As an example, imagine you would like to find 10–15 accessions of beans that are tolerant to high rainfall and low temperatures to include in a breeding program. Searching Genesys for the genus Phaseolus yields about 160,000 accessions conserved at over 97 genebanks. Where do you go from there?
You can further narrow the search down to lima beans (Phaseolus lunatus). That doesn’t help too much: now you have about 8,200 accessions in 37 different genebanks. This is precisely where the Subsetting Tool comes in – when the search is narrowed down, but not quite enough to make a decision on what to select.
Out of those 8,200 lima bean accessions, around 4,200 have the geographical coordinates of their collecting sites included in their passport data. Luckily, each location in the world has a set of climatic and soil data associated with it, thanks to CLIMAP. CLIMAP is an ecographical dataset that provides data on consecutive dry days, total precipitation, average minimum temperature, and so on for all the locations in the world.

Every accession with coordinate data in Genesys is associated with the particular climatic and soil parameters at that location as recorded in CLIMAP.
Using this information, the Subsetting Tool can group your list of accessions according to their similar climate and soil conditions. You can specify which of the variables to consider in order to create a subset relevant to your needs, and also how many subsets the tool should create.

What you cannot control is the number of accessions that this tool yields. Let’s say the set highlighted in yellow above is the one you are interested in: accessions collected from areas with high average temperatures and extremely low average precipitation. This set, however, still contains a lot of accessions: 1,200 of them.
Don’t worry. The Subsetting Tool provides more ways to get to a manageable number, and the means to directly request the accessions from the genebanks through Genesys.

Trying out the Subsetting Tool
The Subsetting Tool is accessible only to registered Genesys users, so make sure to create your Genesys account if you haven’t done so, and be logged in before you test out the tool. Once you are logged in, here’s how you can access it:
-
Navigate to Passport data in Genesys.
-
Specify relevant filters.
-
Apply the selected filters.

💡 Tip: It is important to use passport data filters to reduce the analysis from the over 4 million accessions published in Genesys to a few thousand before running the Subsetting Tool, in order not to fry your computer.
When you are satisfied with the filters you have applied to the accession passport data:
-
Open the Subsetting Tool tab.
-
Expand the categories with the indicators of interest.
-
Select the indicators.

-
Select how many subsets you want to generate.
-
Examine the results in a table.
-
Examine the results through charts.
-
Select the subset that fits your criteria. For example, set 1 below has the highest total precipitation and the lowest number of consecutive dry days.

-
Select a more manageable number of accessions using one of the three options.
-
In the case of a Random Selection or Core Collection, select the total number of accessions to generate.
-
From the generated accessions, select the ones you want to add to My List to request from the genebank(s).

Learning more
The Subsetting Tool offers two modes for selecting indicators: basic and advanced. If you would like to start with the basic mode, you can see the essentials in this short video. Our detailed user guide provides more information on the advanced mode.
We thank the development team at Alliance of Bioversity International & CIAT and ICARDA for their dedication and hard work over many months that resulted in making this tool a reality. This work would not have been possible without the support of the CGIAR Genebank Platform, all contributors to the CGIAR Trust Fund, and all the donors of the Crop Trust Endowment Fund.
7. Order accessions from genebanks through Genesys
Many of the seeds (and other types of planting material) whose data is shared through Genesys are available on request from their holding genebank. When this is the case, Genesys relays requests for material to the genebank. Please keep in mind that most genebanks distribute small amounts for research and breeding purposes, and access is subject to legal and phytosanitary conditions.
Finding accessions
There are many ways to find the crop diversity you are looking for in Genesys. You can conduct a basic search from the Genesys entry page, or an advanced search by filtering passport data, or you may find accessions of interest in subsets and characterization and evaluation datasets.
My List
When you find accessions you wish to request, you need to add them to a list that Genesys calls My List, although it’s actually your list.
You can access My List at any time from the main navigation bar. Here, you can use the Add multiple accessions button to populate your list – you can even paste a list of accession numbers from a spreadsheet.
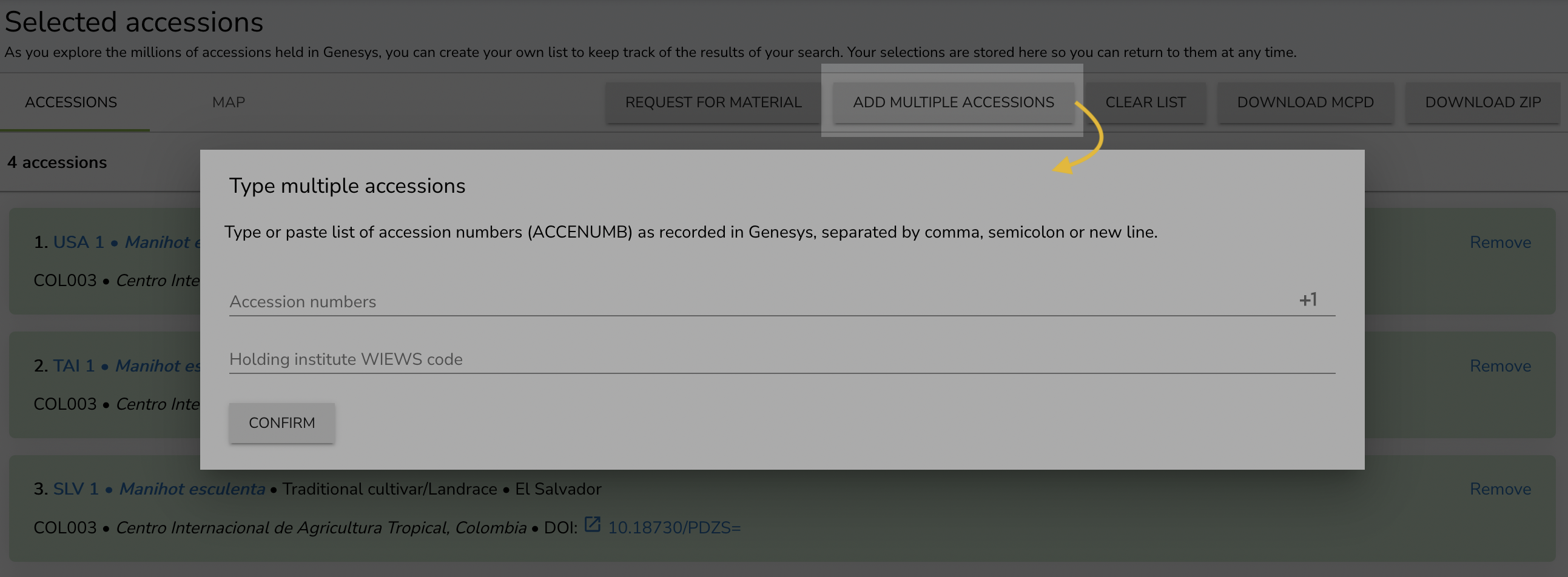
Another approach is to populate your list while browsing Genesys.
Adding accessions to your list as you browse
If you have found an interesting accession in the passport data browser, a subset or trait dataset, or anywhere else, the button you want to look for is + Add to My list on the far right of the accession description.

Alternatively, you can also use the Table view in the Accession browser to have a look at the complete passport data and maybe spot multiple accessions of interest more easily. Select the accessions you want to add to your list by checking the boxes in the My List column.
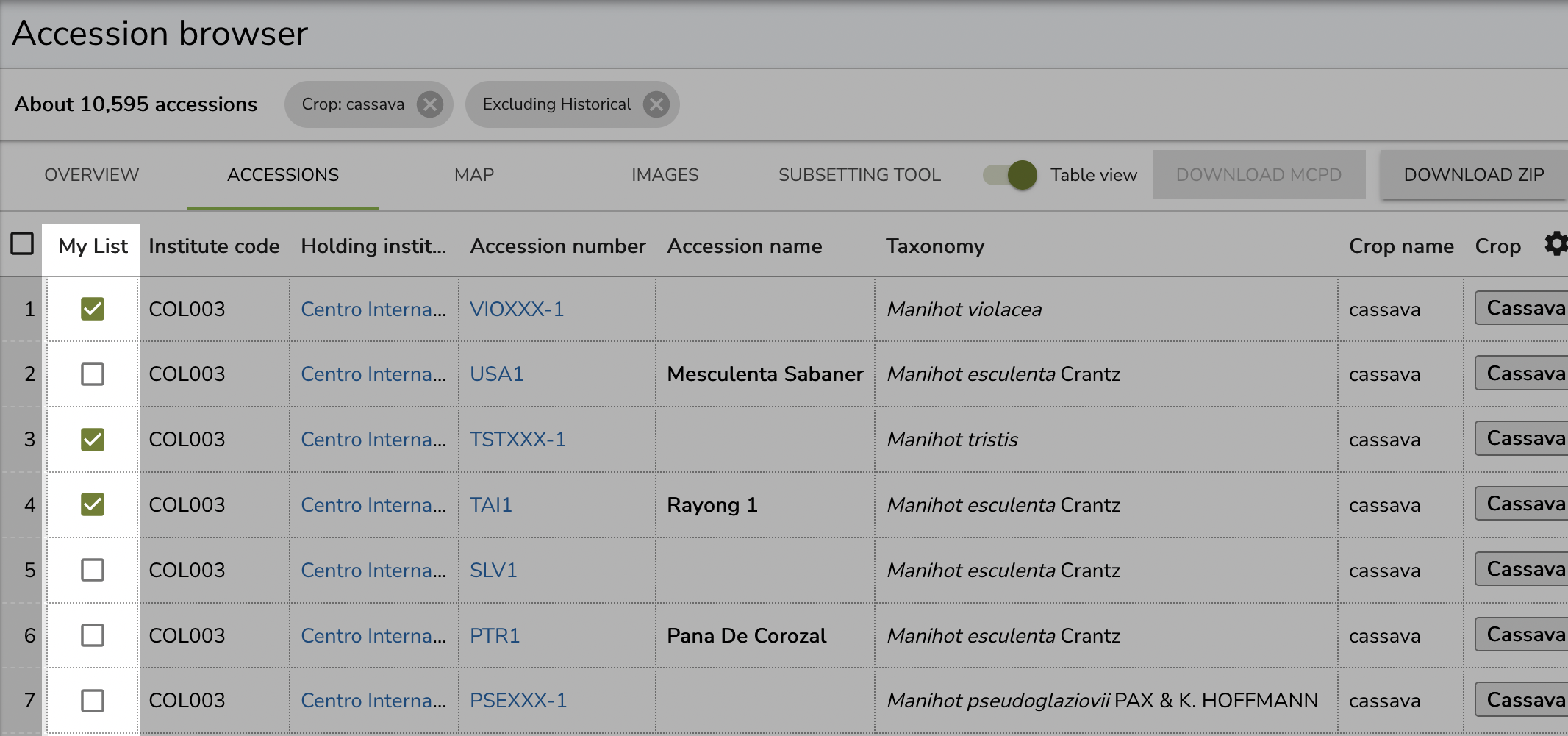
Once you have added all the material that you want to get a hold of to your list, you’re ready to initiate the request process.
Requesting accessions
Now that you have selected the accessions that you need for your research or breeding project, you can send a request to their holding genebank – or genebanks. Genesys allows you to request material held at different genebanks simultaneously.
First, go to My List and select Request for material.

Genesys will first check which of the accessions can be requested. If any accessions are highlighted in red, these cannot be requested through Genesys and they will be automatically excluded from the request. This is the case when the genebank hasn’t enabled the option to receive requests for material though Genesys, or when an accession is currently flagged as “unavailable for distribution”. But don’t panic: you may be able to find a similar accession held in another institute using the Similarity Search function, and add those to your list instead.
Alternatively you can contact the genebank directly. Each genebank page in Genesys includes a Show in FAO WIEWS database link to the respective FAO WIEWS record, where you can find genebank contact information.

For the accessions that are available to request, follow all the steps in the menu box and fill out all required fields. Please flag your request as Internal if you are requesting material from your own institute. Internal requests do not require a material transfer agreement and they are not linked with Easy-SMTA.
Confirming your order
After submitting your request, you will be sent an email with a verification key in order to validate and confirm the order. Please follow all the steps listed in this confirmation email. Only after successfully completing these steps will your request be relayed from Genesys to the holding institute(s).
Patience
Genebank accessions are unlike anything else you order online. Access is subject to legal and phytosanitary conditions, in addition to genebank distribution policies. Most genebanks distribute small amounts, and only for research purposes.
After receiving your request, the genebanks will get in touch with you directly for further details, including import and phytosanitary requirements, material transfer agreement, etc.
Possible claims and complaints related to germplasm requests should be addressed to the specific institute concerned, and not to Genesys, which is just a go-between. You can find the contact information of the holding institutes in the FAO WIEWS database, our data provider directory, or any internet search engine.
8. Watch video tutorials in English
How to Use Genesys
In the webinar How to Use Genesys, we discussed:
The basics
-
Introduction to Genesys 00:00:00
-
Genesys homepage interface 00:03:14
-
Accession-level data in Genesys 00:04:30
-
MCPD descriptors and Genesys 00:07:50
Some useful tools
Browsing and visualization
-
Browsing the database by crop 00:13:37
-
Using the filtering panel and downloading MCPD files 00:15:38
-
Data visualization: Overviews 00:18:22
-
Data visualization: Maps 00:19:24
Filtering and searching passport data
-
Taxonomy and synonym filter 00:21:37
-
Full text search 00:25:33
-
Adding accessions to “My List” and initiating a request for material 00:27:00
-
Historical records in Genesys 00:28:45
-
Different ways to select accessions 00:31:12
-
Geographical filters 00:32:45
-
Filtering by climatic parameters 00:34:53
-
Metadata filters (dates of data upload/update) 00:36:50
-
Genebank profiles 00:37:52
-
Genesys APIs and packages 00:39:13
The session ended with a Q&A session (00:40:30 to 01:15:30) with Matija Obreza, head of genebank information systems at the Crop Trust. Here Matija covered the reuse of data from Genesys, different file formats generated by Genesys, updating and uploading passport data, FIGS subsetting and filtering by climatic parameters, availability of different data types in Genesys, and more.
Subsets and Trait Data in Genesys PGR Database
The webinar Subsets and Trait Data in Genesys PGR Database focused on how to manage collection subsets and trait data in Genesys.
Genebanks use subsets to promote groups of accessions with specific traits or traits of actual or potential interest to users, or that otherwise make sense together, and have sufficient quantity for immediate distribution. Examples include drought-tolerant material, accessions that have been purified and sequenced, and core collections. Users may request the entire subset from the genebank, or pick only some accessions from the list. Of course, users still have the possibility to choose material from the entire collection, or implement their own methods for selecting accessions for their research based on data available to them in Genesys and elsewhere. But having ready-made subsets available can save some time and effort.
Another way Genesys facilitates user access to collections is via characterization and evaluation (C&E) data. Such datasets must be well documented to be published on Genesys. That means having full metadata about how the trait observations were made and how they were recorded. This metadata is crucial for users to be able to make sense of the C&E data and reuse it in their analysis. If the value of a column labeled “RES” for an accession is given as “5” in an Excel spreadsheet, for example, there needs to be an explanation that “RES” refers to level of resistance to a specific disease under specific circumstances, say, and that the “5” was the score on an ordinal scale. Trial conditions, methodology, treatments, and data licensing must be sufficiently documented or users risk making incorrect assumptions and invalid conclusions. Contact information about the owner of the data and links to publications, when available, are very useful in providing context about each dataset. During the webinar, we revisited the key elements of descriptor development (based on Guidelines for the development of crop descriptor lists) and discussed the C&E metadata handled by Genesys.
The slides used in the webinar are also available as a PDF.
9. Watch video tutorials in French
Dans Comment Utiliser Genesys (lien vers l’enregistrement), nous avons abordé les points suivants:
Démarrer dans Genesys
-
Introduction de la présentatrice et de Genesys de 00:00:00 à 00:08:43
-
Types de données dans Genesys de 00:08:43 à 00:10:25
-
Comment utiliser Google Chrome pour traduire automatiquement Genesys en français de 00:10:25 à 00:11:45
-
Se connecter à Genesys et introduction à l’interface de 00:11:45 à 00:18:38
Fonctionnalités, filtres et outils de Genesys
-
Liste des fonctionnalités de Genesys et aperçu de 00:18:38 à 00:21:35
-
Comment utiliser le panneau des filtres des données de passeport - présentation de 00:21:35 à 00:26:47
-
Comment sauvegarder les combinaisons de filtres (URL) de 00:26:47 à 00:28:13
-
Comment utiliser le panneau de filtres des données de passeport - démonstration en direct de 00:28:13 à 00:40:13
-
Comment filtrer par paramètres climatiques et utiliser les superpositions de cartes climatiques - démonstration en direct de 00:40:13 à 00:46:46
-
Questions de 00:46:46 à 00:49:00
Répertoire, téléchargements et demandes de matériel
-
Regroupement des accessions dans le répertoire de Genesys - présentation de 00:49:00 à 00:52:42
-
Téléchargement de fichiers de données de passeport et standard MCPD - présentation de 00:52:42 à 1:06:15
-
Démonstration en direct du répertoire et du PDCI (Indice de Complétude de Donnees de Passeport) de 1:06:15 à 1:11:09
-
Questions de 1:11:09 à 1:15:00
-
Outils utiles de la banque de gènes (montrer les changements, recherche de similarité) de 1:15:00 à 1:31:00
-
Demande de matériel par le biais de Genesys de 1:31:00 à 1:33:10
Questions et réponses de 1:33:10 à 1:39:03